Research
I am currently a PhD student in Electrical and Computer Engineering specializing in computational biology. I am particularly interested in applications of artificial intelligence (AI)/machine learning (ML) to design novel therapeutic peptides and study lysine methylation. I have also developed an interest in high performance computing throughout the past years. My thesis advisors are Profs. James R. Green (Systems and Computer Engineering) and Kyle K. Biggar (Biochemistry).
Current research
As part of my thesis work, I apply AI/ML tools to the study of lysine methylation. My research can be subdivided into three research themes:
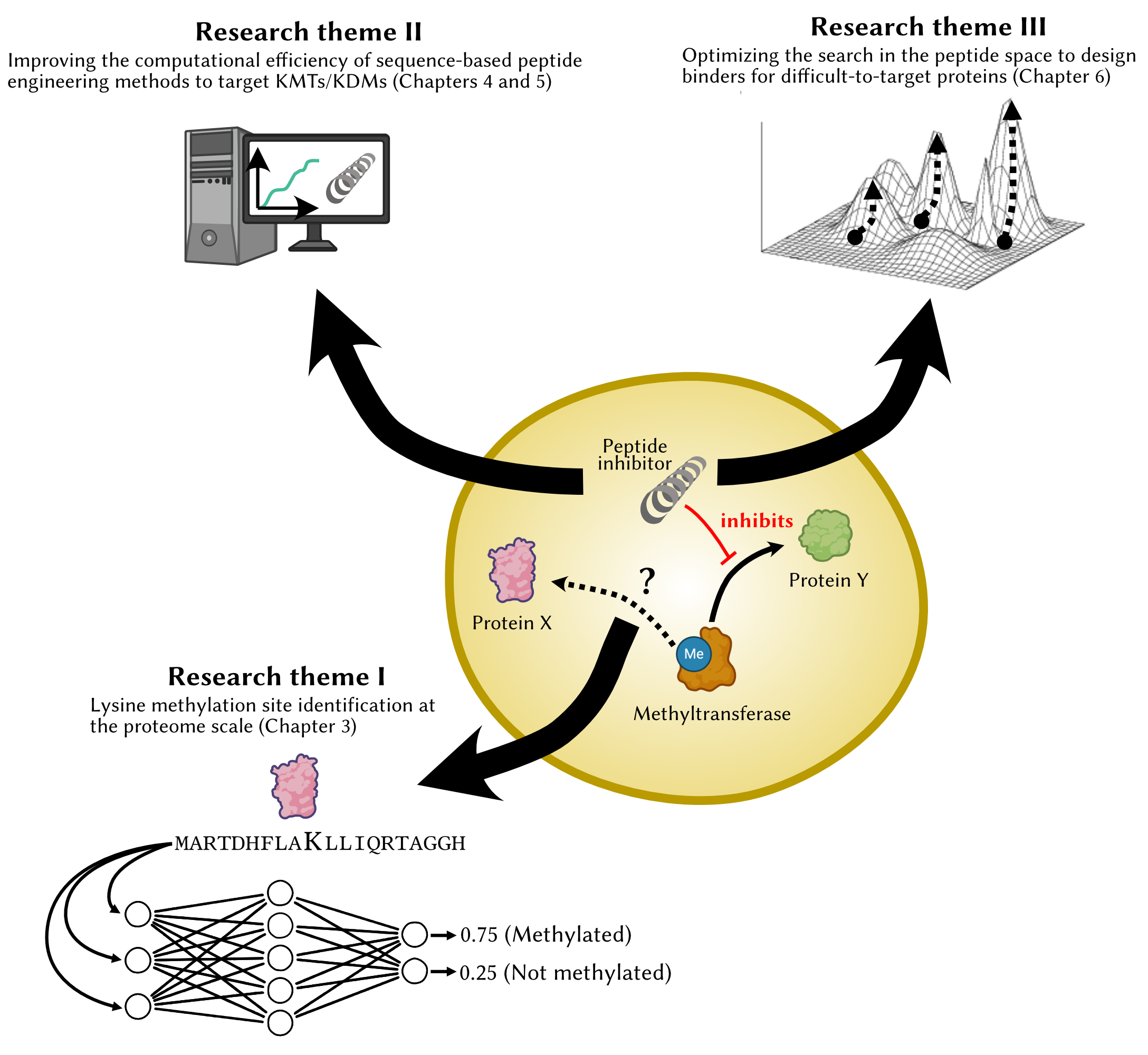
- Proteome-wide identification of the lysine methylation sites
Our cells express over 20,000 proteins, a large portion of which are substrates in a set of chemical reactions called post-translational modifications (phosphorylation, acetylation, methylation, etc.) These reactions can modulate the function, stability and localization within the cell. I aim to develop new machine learning approaches to answer questions such as: What proteins are methylated? How does this relate to other modifications? What are the implications of this modification? - Improving the computational efficiency of in silico design of lysine
methyltransferase inhibitors
I am improving upon the state-of-the-art algorithms for peptide inhibitor design using sequence information only, as existing algorithms are computationally intensive and not scalable. We are developing a fast algorithm named Darwin which leverages large databases of protein-protein interactions active peptides capable of specifically modulating the activity of lysine methyltransferases (KMTs) and lysine demethylases (KDMs). Overactive KMTs/KDMs are involved in a number of cancers, and these peptides could supplement existing therapies and improve their efficacy. - Optimizing the design process for difficult-to-target proteins
Sequence-based peptide engineering methods rely on validated protein-protein interactions (PPIs). In some cases, databases of known PPIs provide insightful information that can guide the peptide design process. In other cases, the finite set of known interactions is insufficient as the target may bear little similarity to proteins with known interactors. I am trying to improve the search strategy in a very large peptide space in order to probe it more efficiently using deep learning techniques (protein language models).
Want to know more about my main research project? Feel free to take a look at my submission for the 2021 Gradflix contest:
Previous research
- Structural biology
Under the supervision of Prof. Jean-François Couture, I completed my my honours thesis on the structural characterisation of the Fur (Ferric Uptake Regulator) protein in Campylobacter jejuni a pathogenic agent responsible for numerous cases of gastroenteritis. I crystallized the protein and built a draft model of the structure, which was further refined by Prof. Couture. This research was published in the scientific journal Scientific Reports. - Computational audiology
I have previously worked in computational audiology. My master's thesis work focused on the ML-assisted classification of audiograms to facilitate their interpretation by non-experts.